New imaging tool for recording changes over time, space and function
Researchers who work with stem cells have ambitious goals. Some want to cure cancer or treat heart disease. Others want to grow the tissues and organs that patients need for transplants. Some groups are even working to develop highly personalized medicines, tailored to an individual’s genetics. All of these ideas face a similar hurdle, however: The development of measurement tools for stem cell production is challenging, making it hard to determine what makes various new stem cell-related products safe, effective or high-quality.
Over the last few years, NIST scientists have been trying to solve this problem in a way that will seem familiar to anyone who has ever used a smart phone and a mapping application. The system they have developed, however, goes way beyond anything found in a typical app. It combines video footage and high-power computation to bring the world one step closer to evaluating, understanding and quantifying features of cell populations that could be used in related therapies or products.
The new image analysis system, called Web Image Processing Pipeline (WIPP), is like a mapping app that allows a user to interact with microscopic views of macroscopic objects. With WIPP, a cell culture is divided into real locations that can be explored and referenced with a system very much like GPS points or the quadrants on a geographic map or mapping app. But WIPP also allows a user to examine what’s happening in a petri dish over time, from any point of view they choose.
The work on this new interface is detailed in a new reference book, Web Microanalysis of Big Image Data, published in January 2018 by Springer Verlag. It was also described in an article published in the journal Stem Cell Research in May 2016.
“You can share results with colleagues and analyze what’s happening with these cells in many different ways—in space, over time and by their function. And you can review how the cell changes in time, over and over again,” said Peter Bajcsy, author of the book and the IT lead for the research behind WIPP.
WIPP was designed for work with induced pluripotent stem (iPS) cells, which are cells taken from the tissue of an adult and processed to behave like an embryonic stem cell. iPS cells can be used to make new cells from all three of the basic human body layers: ectoderm (skin), endoderm (gastrointestinal and respiratory tracts), and mesoderm (blood and bone). iPS cell technologies are still relatively new, and it is unclear which tissue types derived from iPS cells will have the greatest impact first. Researchers are seeking ways to effectively and reliably measure progress as cells become different cell types during processing.
Model iPS lines are usually purchased from established cell banks, which operate somewhat like blood banks. Depending on the application and the laboratory in which they are grown, the cells are cultured in different specialized media and on different specially treated surfaces. The media are replenished on a regular basis to keep the living cells in a pluripotent state as they expand in number. They can be used in experiments to study fundamental biology, or they can be differentiated into specific mature cells that can be developed for medical treatments or used to test drugs.
Every cell matters. If one undesirable cell reproduces rapidly and comes to dominate the culture, it could become a tumor or make the preparation otherwise unsuitable for use as a therapy. To fully understand what’s happening in these stem cell systems, a researcher would want to know the characteristics of the cells in the dish by watching them change, grow, reproduce or die in order to eliminate as many of the potential dangers for patients as possible.
“We believe live cell imaging is really the only tool you can use to follow single cells in time and understand at the single cell level how behavior at a given point in time is related to the future,” says Michael Halter, team leader for image acquisition for the WIPP project. “We don’t just want to analyze one colony or a few colonies. We want to look at all of the colonies and we want to look at them over time.”
How WIPP Works
At first, the entire concept of live cell imaging research seems simple: take footage of given cell colonies using existing, well-known tools like video microscopy. Run footage and observe what happens to the cells over time.
The problem, however, is one of scale and size. Each dish is full of thousands of cells, and taking enough images to capture movement and changes in each of those cells can demand a lot from a computer. For instance, three runs of a 5-day experiment can yield a data set equal to a terabyte (1 TB = 1012 bytes). Trying to transfer data onto the hard drive of a laptop or desktop system—loading it into a computer memory that is often only 1-3 terabytes in total size—and working with it in any sort of nimble or flexible manner is impossible because of current hardware and software limitations. Without adequate processing and computer memory, those laptops and smaller desktops begin to choke.
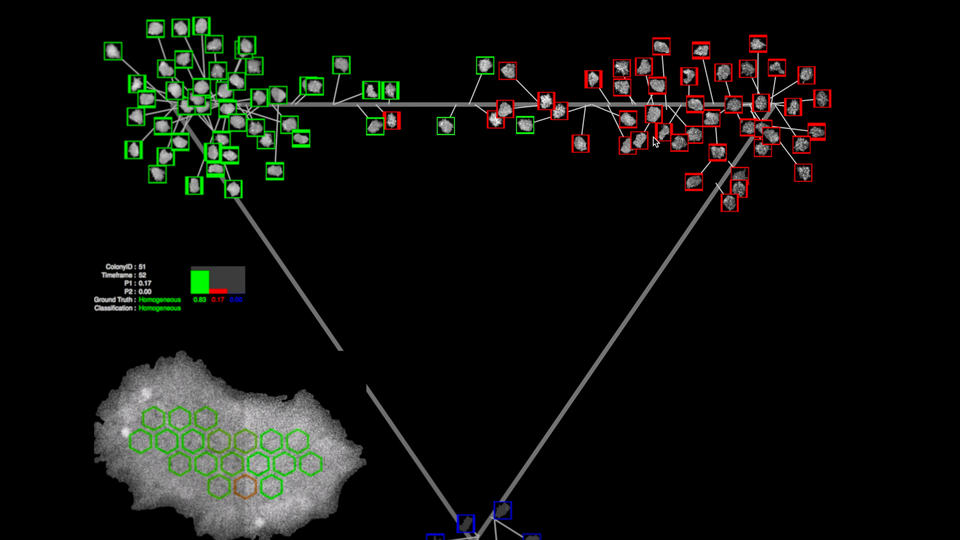
A still image from the WIPP software, which shows several cells outlined in red and several in green against a dark background. Also pictured: a map of the cells geographic location within the petri dish. Credit: Jose Garcia/NIST
WIPP circumvents this problem by leveraging the computational power of a server and the flexibility of a web interface. Researchers upload their data to the server, which carries out some computational operations, while others are carried out on a desktop with the web browser. The entire WIPP system is open access and in the public domain, although researchers’ own datasets remain private for specific projects until they choose to make them public.
Sections of the cell culture dish where a colony is being grown are photographed in sequence with use of a microscope. Those images are then stitched together, like a quilt, to form a view of the entire dish at each point in time. Each of those complex composite, single-time-point images are then run like a movie, revealing the cells’ entire history.
Although it sounds simple enough in concept, the design and modeling process proved to be enormously difficult and computationally challenging. First, experts from biology and computing domains had to find a common language in order to match biological knowledge to mathematical models and information technologies. Next, the new process had to be validated so that any resulting measurements could be trusted by users. The measurements then had to be integrated into one product.
Finally, the complex algorithms used for image calibration had to be designed to run efficiently—not only on the system’s servers, but also on potential client computers, using many types of internet browsers. These algorithms addressed complex needs such as: background and flat-field correction, stitching to make those “quilt-like” arrangements of images, the segmentation of cell colonies or cells, and feature extraction, in order to characterize segmented objects. The result feels a lot like an online Earth-mapping app accessed via a phone. Users can start with a view of a very large ‘map’ of the petri dish that shows all the cells and use a cursor to jump from one part of the dish to another, zooming in and out for closer looks as needed. They can switch from a coarse level of detail to a finer one. They can also stop to examine the images at precise moments in time and move forward or backward over seconds, minutes or days to see the entire life span of a cell.
The key difference between regular online mapping and WIPP is the amount of complexity built into the system. When you use your cell phone to find a place like a city park via an online map, you are looking to see where the park is and how to travel from your location to get there. In the WIPP system, you can travel around in various maps, and visit different places in the petri dish of cells. But you can also see what happens in each of those locations over time and how the cells there change and grow. It’s almost like using your cell phone to see where a park is located, but also the age of every squirrel, bird and worm living there, the number of offspring they’ve had, and the amount of food they ate at a specific time on the morning you visited.
WIPP incorporates the computations behind the images, pulling in the analytical data to match each view of the experiment. This allows users to collect the key measurements about shape, size, texture features, fluorescence intensities, contrast and rates of change in these measurements, and to analyze the cell population within each colony over time and space.
You can also keep that computational data and the associated images and remix them over time in different ways. If you want to see whether one cell lived or died, for example, you can do that. But if you decide later that you want to see which cells reproduced rapidly and which ones reproduced slowly during your previous experiment, you can do that, too.
Researchers can even use the WIPP system to gain traceability of computations in their experiments, which has become particularly important over the last few years as several high-profile studies have been retracted when it was discovered that their results could not be reproduced in follow-up experiments.

Micheal Halter (on left) and Peter Bacjsy, during a live demonstration of the software. Credit: Jose Garcia/NIST
“If somebody doesn’t trust your numbers,” Bajcsy says, “they can use this tool to go back to your raw field of view and the original set of images. For traceability of computations and reproducibility of experimental results, that’s really important. And you cannot trace results if some computation is done on this desktop and some on another laptop and the algorithmic parameters, software versions, software configurations and chains of computations are not recorded and linked automatically. Now you have one central repository with everything.”
The system could also be used to evaluate next-generation pharmaceutical products during and after manufacturing. Quality assurance has long been a part of pharmaceutical manufacturing, but it is becoming a more complex issue as the use of live cells as therapies becomes more common. The recent Food and Drug Administration approval of a genetic therapy for leukemia provides one case in point. In such manufacturing processes, the assessments and measurements of quality must be reproducible and precise because lives are at stake. That kind of quality control must be done at the cell population level, which demands the sort of large data scales that WIPP can process.
A Unique Collaboration
WIPP was the result of a collaborative effort between two teams of NIST experts: those working to support high-quality measurements of cellular systems and those working in information technology.
“If we had done this by ourselves, we would have gotten an off-the-shelf software tool kit and never gone this direction,” says Halter, a biologist. “Now a powerful software system is available to a wide range of researchers.”
“Without working together and combining the biological and modeling expertise, we cannot obtain meaningful analyses of these very large imaging experiments,” Bajcsy says.
NIST is offering use of the WIPP interface to anyone who wants to use it for research purposes, free of charge. The open-source code used to create it is also available. Halter and Bajcsy hope that others who are working to further expand the potential of stem cells will expand or improve upon WIPP within new applications.
“The biology of the body is so complex,” says Halter. “We need sophisticated approaches to evaluate new products from stem cell research before they are launched in medical devices or in people. This tool is a really important step in that direction.”
Source: NIST
