Scientists at Colorado State University (CSU) and Tokyo Institute of Technology (Tokyo Tech) have developed a new powerful tool called a “frankenbody”, for imaging protein dynamics in living cells. Just as Frankenstein's monster is stitched together from different limbs, the frankenbody was created by taking the binding regions of a normal antibody, the “sticky parts,” and grafting them onto a different scaffold that remains stable in live cells but retains the specificity of the antibody.
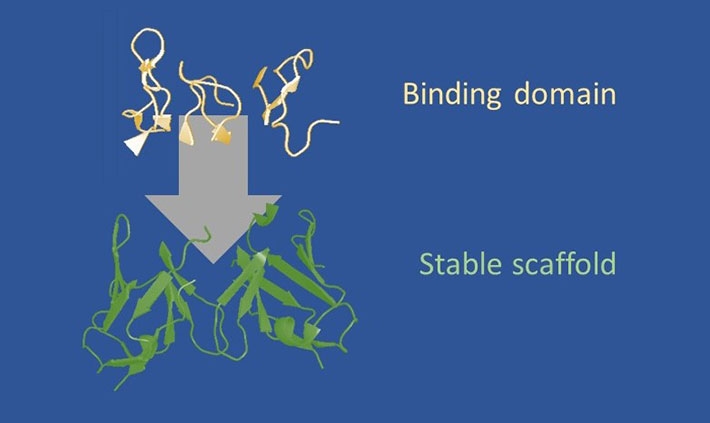
Figure 1. Design strategy of frankenbodies. Image credit: Tokyo Institute of Technology
A diverse set of scFv scaffolds proven to fold and function in living cells. Onto these scaffolds, we loop graft all complementarity determining regions (CDRs, or loops) from an epitope-specific antibody. Depending on the compatibility of the scaffold and CDRs, this produces a hybrid scFv that retains the stability of the scaffold, while acquiring the specificity of the grafted CDRs.
Antibodies are the biomolecules our immune systems deploy to find, tag, and destroy invading pathogens. They work by binding to specific targets, called epitopes, on the surfaces of antigens – like locks to keys.
For many decades, scientists have cleverly exploited this selective tagging mechanism in natural antibodies to engineer antibody-based probes that let them purify and study different types of proteins within cells. One tried and true technique, epitope tagging, involves fusing an epitope to a protein of interest and using fluorescently labeled antibodies to make those proteins visible – but only in fixed, dead cells.
A useful complement to the green fluorescent protein
Now, a cross-disciplinary team of researchers from Tokyo Tech and CSU have added a new tool to the arsenal of antibody-based probes, but with a powerful distinction: their genetically encoded probe works in living cells. The work, led by CSU Monfort Professor Tim Stasevich and Tokyo Tech Professor Hiroshi Kimura, was published on July 3rd, 2019 in the journal Nature Communications.
They engineered the frankenbody as a hybrid of single-chain variable fragments that retains the stability of the scaffold and the specificity of the grafted complementarity determining regions (Fig.1). “We're interested in intracellular antibodies because you can use them as imaging reagents in a live cell,” said Stasevich, an assistant professor in the Department of Biochemistry and Molecular Biology at CSU. “You don't need a tag, like a green fluorescent protein (GFP), because instead you have this fluorescent antibody that will bind to your protein that you want to visualize.”
The new probe would be a useful complement to the GFP, a widespread biochemistry tool and Nobel prize winner that involves genetically fusing a light-up green tag to a protein of interest. However, the GFP is limited by its relatively large size and the time it takes to fluoresce; with the new probe, the tag is smaller and becomes fluorescent faster, so the “birth” of a protein of interest can be captured in real time.
With the goal of making their tool immediately useful, the scientists designed their probe to work with the classic HA tag. HA is a widely used small linear epitope tag that's derived from a portion of the human influenza virus protein hemagglutinin (Fig.2).
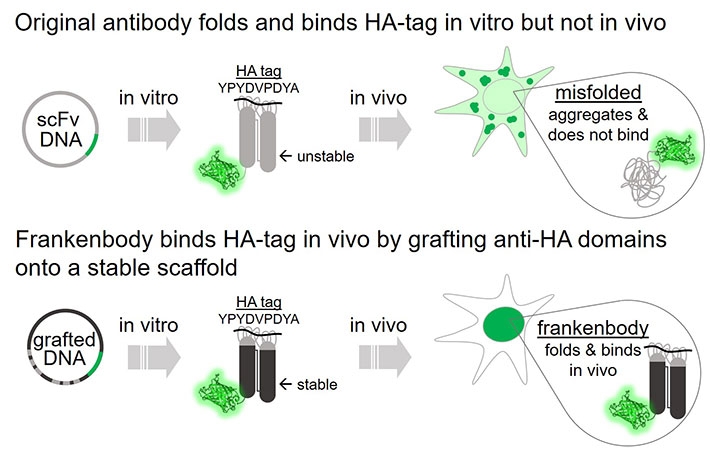
Figure 2. Frankenbody binds HA-tag in vivo. Image credit: Tokyo Institute of Technology
This wildtype anti-HA scFv does not fold properly in the reduced intracellular environment, and therefore displays little affinity for HA epitopes in living cells. However, both nuclear and cytoplasmic HA-tagged proteins can be labeled by the HA frankenbody in living cells.
“For the longest time, people have been looking at HA-tagged proteins in fixed, dead cells,” Stasevich explained. “Now we can image the dynamics of those proteins in live cells.”
A limitless probe
The possibilities of how scientists may use the new probe are limitless. Stasevich's lab is particularly interested in studying RNA translation, and they plan to use their new system to more easily design new RNA imaging experiments.
The frankenbody is genetically encoded on a plasmid that can be easily distributed to other labs. This is in contrast to traditional antibodies, which can cost a lab several hundreds of dollars per order and suffer from lot-to-lot variability. The new probe from Stasevich's team therefore provides a lost-cost solution for protein and RNA translation imaging.
In the paper, the scientists demonstrated applications, including single-protein tracking, single-RNA translation imaging, and amplified fluorescence imaging in zebrafish embryos, all of which pose challenges for more traditional fluorescent protein tags.
The work strengthens ties between CSU and the Tokyo Tech. “We have several new imaging reagents in the works that build off of this success, so I see great things ahead,” said Stasevich.
Reference
Ning Zhao1, Kouta Kamijo2, Philip D. Fox1, Haruka Oda2, Tatsuya Morisaki1, Yuko Sato2, Hiroshi Kimura2, and Timothy J. Stasevich1,2. A genetically encoded probe for imaging nascent and mature HA-tagged proteins in vivo. Nature communications . DOI: 10.1038/s41467-019-10846-1
1Colorado State University, USA
2Tokyo Institute of Technology, Japan
Source: Tokyo Institute of Technology
