Cold Spring Harbor Laboratory (CSHL) Associate Professor Pavel Osten and his lab map brain connections and catalog different cell types throughout the brain in a quantitative brain-wide (qBrain) catalog. qBrain combines several techniques developed by CSHL and other scientists to label, preserve, image, visualize, count automatically, and trace cells.
With these methods, the researchers can standardize a 3D map of the 100 million neurons in the mouse brain and in the near future, even the 100 billion neurons in the human brain. The researchers can compare new brains to their standardized 3D maps to figure out gender differences within a species, development stages, and diseases.
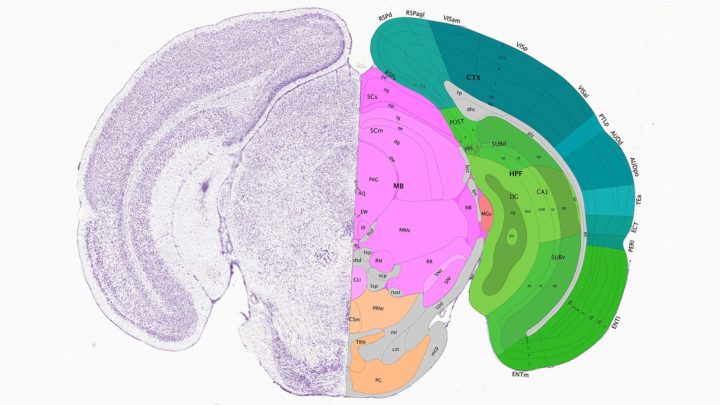
A slice of a mouse brain, with a detailed microscopic image showing individual neurons on the left and a map showing the labeled structures in the Common Coordinate Framework on the right. Image credit: Allen Brain Atlas, atlas.brain-map.org
How qBrain images and analyzes brains
Osten’s technique starts with labeling brain cells of interest to identify classes of cells or particular pathways. The brain is then preserved and imaged automatically at high resolution. Each brain is analyzed by a computer, which can count the cells or trace the pathways, comparing it to previously mapped brains. The entire process, automated after the brain is preserved, takes 12 to 32 hours per mouse.
qBrain builds a standardized quantitative brain map by not only locating pathways but counting the number of cells that participate in that pathway. For any particular species, as each new brain comes in for mapping, it is added to the previous data to register (fit) it to a standard “average” brain. Osten explains:
“We take the average brain and stretch it or squish it onto each of the new brains. It’s really essential that you have everything in the same space because each brain can be a little bit different, bigger or smaller. So in order to compare between brain regions, the first step is to be able to register all the data in the same space.”
Once the researchers determine a standard brain, they can then measure variations between individuals of the same species and between the same brain regions in different species.
One qBrain application: defining gender differences
Osten and his collaborators compared average male and female mouse brains. The female mouse brain has more complex cell type distributions in regions that are involved in parenting and nest building. The researchers believe the richer cell type compositions are associated with pup-rearing, which females do; males are not involved.
Scaling up to more complex brains
Osten and his lab were a part of the founding group of scientists for the NIH-funded Brain Research through Advancing Innovative Neurotechnologies (BRAIN) Initiative – Cell Census Network (BICCN). Similar in scope to the Human Genome Project, BRAIN is providing a foundation for studying the mammalian brain with new methods. Osten says that they have come a long way in the last few years. “The first three years were all about developing the methods and testing them. Over the last five years, we applied the methods mainly to the mouse. Next, we are hoping to apply these methods to the human brain.” Osten believes that over the next few years it will become possible to image and analyze an entire human brain in only a few days.
Osten is working with many collaborators to scale up and study whole human brains, including Elizabeth Hillman, a professor of biomedical engineering and radiology at Columbia University, and Zhuhao Wu, an assistant professor at Mount Sinai University. Osten speculates:
“Doing one human brain would be a success, but not really very much of an advantage because you have to know what’s the variability between different humans, what’s the variability in disease. So part of the method that we are building with Elizabeth and Zhuhao is to be really able to do tens of thousands of human brains.”
Osten’s technological advances and findings will help researchers better navigate the complex human brain, yielding deeper understandings of all the cells, connections, and differences that make us human.
Source: CSHL
